Introduction to Lineweaver-Burk Transformation
July 29, 2020 ☼ Molecular Biology
In the last post, I introduced one of the fundamental models for enzyme kinetics, the Michaelis-Menten model. While the Michaelis-Menten Model serves as the foundation for modeling enzymes, there have been additional transformations and modifications to make the model more tractable for experiment-based characterization. One of these approaches, the Lineweaver-Burk Transformation, is a simple mathematical reorganization of the Michaelis-Menten Model that enables deeper interrogation enzyme mechanisms or inhibition patterns through graphical analysis. Let’s see how this works!
Table of Contents
Model Background
The Lineweaver-Burk transformation is a mathematical reorganization of the original Michaelis-Menten first established by Hans Lineweaver and Dean Burk1. Because it is just a transformation of the original equations, it is subject to the same assumptions and restrictions as the original model. Recall that the Michaelis-Menten equation is:
$$v = \frac{V_{max} [S]}{K_m + [S]} \tag{1}$$
Where:
- $v$: Steady-state reaction velocity
- $V_{max}$: Maximal reaction velocity when all enzymes are saturated with substrate
- $[S]$: Concentration of substrate
- $K_m$: Michaelis-Menten Constant
To generate the Lineweaver-Burk transformation, you take the reciprocal of both sides of the Michaelis-Menten equation:
$$\frac{1}{v} = \frac{1}{V_{max}} + \frac{K_m}{V_{max}}\frac{1}{[S]} \tag{2}$$
The Lineweaver-Burke transformation changes the form of the Michaelis-Menten equation into something that should be familiar: the general form for a line. In this case:
$$\text{Y-intercept}: \frac{1}{V_{max}} \text{ | } \text{Slope}: \frac{K_m}{V_{max}}$$
Model Analysis
Let’s compare the graphs of the MM equation and the LB equation under the same conditions.
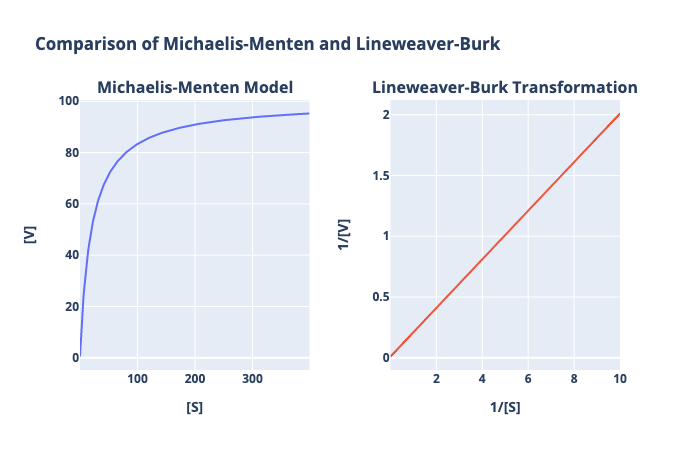
Because the Lineweaver-Burke plot is a linear transformation, it’s much easier to do a graphical analysis for enzyme kinetic constants. For example, instead of extrapolating out the asymptote in the Michaelis-Menten plot to determine $V_{max}$ (remember, $V_{max}$ is not always known a priori), we know that the reciprocal $V_{max}$ is simply the Y-intercept of the Lineweaver-Burk plot.
What about $K_m$? Identifying and approximating the $K_m$ appears to be easier in the Michaelis-Menten plot since it works directly with the substrate concentration. However, identifying $K_m$ is the Lineweaver-Burk plot is also relatively straightforward and is even easier to extrapolate. Let’s start with the mathematical definition:
Unlike the Michaelis-Menten case, where we set $v = \frac{V_{max}}{2}$, we are going to set $\frac{1}{v} = 0$:
$$0 = \frac{1}{V_{max}} + \frac{K_m}{V_{max}}\frac{1}{[S]}$$
Rearranging this equation, we get the final mathematical relationship needed to identify the $K_m$:
$$K_m = -1 \div \frac{1}{[S]} \tag{3}$$
This definition may look weird at first, but it has an important graphical meaning. Since we set $\frac{1}{v} = 0$ to reach this point, $\frac{1}{[s]}$ represents the x-intercept of the Lineweaver-Burk plot. In other words:
$$K_m = - \frac{1}{\text{X-intercept}} \tag{4}$$
Further Applications of Lineweaver-Burk Plots
Lineweaver-Burk plots are also used for three additional applications for understanding enzyme kinetics:
- Distinguishing an enzyme’s binding mode (e.g., the order of substrate binding)
- Identifying an inhibitor’s mechanism of action
- Extrapolating experimental data to determine $K_m$ and $V_{max}$
While Michaelis-Menten plots are still valid in each of these applications, the Lineweaver-Burk transformation’s linear nature is more interpretable and easy to use. I’ll dive into each of these applications in future posts.
Interactive Exploration
Because the value of using Lineweaver-Burk plots in mainly graphical, the best way to understand how they work is graph examples yourself. Below, there are links to a Python notebook that provides an environment to explore the model and graphical tools in more depth.
Closing
While the Michaelis-Menten model provided the foundations for understanding enzyme kinetics, the original equation’s transformations have expanded its applicability. The Lineweaver-Burk transformation is a reciprocal transformation resulting in a linearized form of the equation. I hope this brief introduction to the Lineweaver-Burk transformation is helpful and hope you come back for follow-up posts on its applications to characterize enzymes.
Lineweaver, Hans, and Dean Burk. “The Determination of Enzyme Dissociation Constants.” Journal of the American Chemical Society, vol. 56, no. 3, Mar. 1934, pp. 658–66. DOI.org (Crossref), doi:10.1021/ja01318a036.↩