The Three T’s of Gene Transfer
July 8, 2020 ☼ Molecular Biology ☼ Experimental Methods
Ever since recombinant DNA technology transformed basic science and industrial research and development in the life sciences in the 1970s and 1980s, transferring DNA between organisms became a fundamental part of everyday research workflows1. Understanding the basis for these techniques is essential for preparing and planning out many life sciences research projects.
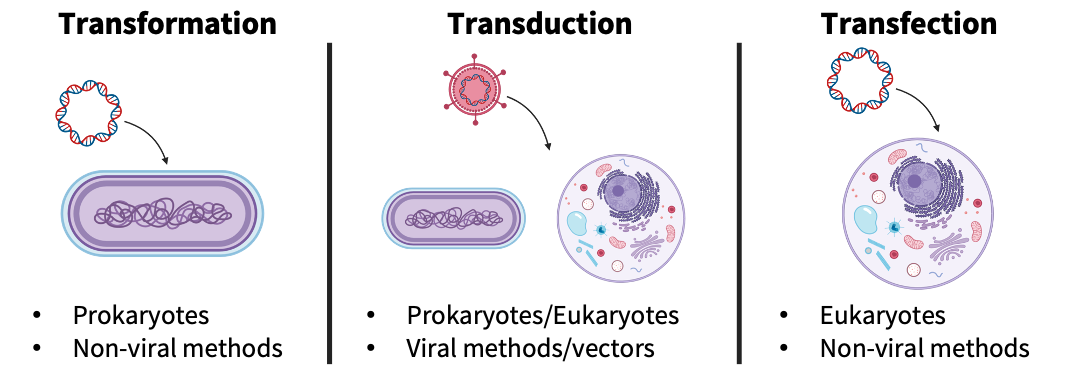
There are three main categories of gene transfer techniques (hence: the Three T’s of Gene Transfer): (1) transformation, (2) transduction, and (3) transfection. These three categories are differentiated based on the target cell type (prokaryotic versus eukaryotic cells) and transfer method (non-viral or viral methods).
 The Three T’s of Gene Transfer
The Three T’s of Gene Transfer
Transformation
Transformation is the process of transferring DNA into prokaryotic cells (e.g., E. coli, yeast) through non-viral methods. This technique is based on bacteria’s ability to uptake genetic material from the environment around them. Not all bacteria can uptake genetic material or have low uptake rates. To improve the uptake rate (also known as the transformation efficiency in a laboratory setting), scientists have developed a two-step process:
- Treat cells with chemicals (such as calcium chloride) to mask the cell membrane with positive charges.
- This enables the negatively charged DNA to approach and “stick” to cell membrane instead of being repelled
- You can buy chemically competent cells that are pretreated and skip this step.
- Improve membrane permeability and drive the DNA into the cell using electroporation (apply electric current) or heat shock (apply higher temperature).
In most cases, you will be introducing a plasmid, or a circular piece of DNA, into the cell. This plasmid contains multiple components, including a signal to continually reproduce the plasmid, preventing it from diluting over time through cell growth and division.
Transduction
Transduction is the process of transferring DNA into prokaryotic or eukaryotic cells through viral methods. Using this method, you can stably integrate the DNA into the bacterial or mammalian cell genome, ensuring the new genes are continually replicated. For prokaryotic cells, you’ll often use tools like bacteriophages or phagemids. For eukaryotic cells, you will use tools, like lentiviruses or Adeno Associated Viruses (AAV) that have been modified to reduce potential risks, but still require special safety precautions.
This process is significantly more complicated (and potentially more dangerous if you’re using mammalian cells) compared to the two other methods introduced in this post. Unlike transformation, which only gets the DNA into the cell, transduction integrates the DNA directly into the genome of the cell. While stable integration is not necessary for bacterial cells (as discussed in the section above), stable integration into eukaryotic cells is necessary because these types of cells do not have the machinery to reproduce DNA that are not integrated.
Transfection
Transfection is the process of transferring DNA into eukaryotic cells (e.g., mammalian cell lines) through non-viral methods. Like transformation for prokaryotic cells, transfection is a two-step process consisting of chemically treating cells and using electroporation or heat shock to drive in the DNA. This DNA will be introduced into the cell and can take advantage of the transcription and translation machinery, but is not replicated. This lack of replication results in a dilution and eventual loss of the DNA over time as the cells grow and divide.
Transformation, transduction, and transfection are the three main categories of techniques to transfer genes into cells. These techniques have many things in common, but can be distinguished by the cell type and transfer method. While hands-on experience is essential for applying these techniques in practice, understanding the conceptual basis for these techniques and how they differ can help determine which route is best for your project.
Alberts, Bruce, Alexander Johnson, Julian Lewis, Martin Raff, Keith Roberts, and Peter Walter. 2002. “Isolating, Cloning, and Sequencing DNA.” Molecular Biology of the Cell. 4th Edition. https://www.ncbi.nlm.nih.gov/books/NBK26837/.↩